Mixed logit with ARD
Contents
Mixed logit with ARD¶
We begin by performing the necessary imports:
import os
os.environ["CUDA_VISIBLE_DEVICES"]="0"
import sys
sys.path.insert(0, "/home/rodr/code/amortized-mxl-dev/release")
import logging
import time
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
# Fix random seed for reproducibility
np.random.seed(42)
Generate simulated data¶
For this demo, we generate simulated choice data, but we will set one of the fixed effects parameters and one of the random effects parameters to zero, such that their associatied attributes are irrelevant for the utilities.
Other that that, the simulated data is generated in a similar way to the “Mixed logit with simulated data” demo. We predefine the fixed effects parameters (true_alpha) and random effects parameters (true_beta), as well as the covariance matrix (true_Omega), and sample simulated choice data for 2000 respondents (num_resp), each with 5 choice situations (num_menus). The number of choice alternatives is set to 5.
from core.dcm_fakedata import generate_fake_data_wide
num_resp = 2000
num_menus = 5
num_alternatives = 5
# one of the parameters is set to zero such that the corresponding input variables
# is not correlated with the observed choices
true_alpha = np.array([-0.8, 0.8, 0])
true_beta = np.array([-0.8, 0.8, 1.0, -0.8, 0])
# dynamic version of generating Omega
corr = 0.8
scale_factor = 1.0
true_Omega = corr*np.ones((len(true_beta),len(true_beta))) # off-diagonal values of cov matrix
true_Omega[np.arange(len(true_beta)), np.arange(len(true_beta))] = 1.0 # diagonal values of cov matrix
true_Omega *= scale_factor
df = generate_fake_data_wide(num_resp, num_menus, num_alternatives, true_alpha, true_beta, true_Omega)
df.head()
Generating fake data...
Error: 42.18
| ALT1_XF1 | ALT1_XF2 | ALT1_XF3 | ALT1_XR1 | ALT1_XR2 | ALT1_XR3 | ALT1_XR4 | ALT1_XR5 | ALT2_XF1 | ALT2_XF2 | ... | ALT5_XR1 | ALT5_XR2 | ALT5_XR3 | ALT5_XR4 | ALT5_XR5 | choice | indID | menuID | obsID | ones | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.374540 | 0.950714 | 0.731994 | 0.069212 | 0.585697 | 0.798869 | 0.764473 | 0.837396 | 0.598658 | 0.156019 | ... | 0.119691 | 0.938057 | 0.510089 | 0.300522 | 0.624496 | 0 | 0 | 0 | 0 | 1 |
| 1 | 0.183405 | 0.304242 | 0.524756 | 0.257912 | 0.040451 | 0.793656 | 0.995865 | 0.956444 | 0.431945 | 0.291229 | ... | 0.107102 | 0.495835 | 0.287860 | 0.651430 | 0.145982 | 2 | 0 | 1 | 1 | 1 |
| 2 | 0.607545 | 0.170524 | 0.065052 | 0.588614 | 0.571589 | 0.402953 | 0.482491 | 0.146977 | 0.948886 | 0.965632 | ... | 0.685591 | 0.543384 | 0.531436 | 0.551352 | 0.486627 | 2 | 0 | 2 | 2 | 1 |
| 3 | 0.662522 | 0.311711 | 0.520068 | 0.946478 | 0.618740 | 0.459393 | 0.860055 | 0.889657 | 0.546710 | 0.184854 | ... | 0.071816 | 0.562382 | 0.314172 | 0.099657 | 0.190283 | 3 | 0 | 3 | 3 | 1 |
| 4 | 0.388677 | 0.271349 | 0.828738 | 0.790511 | 0.806431 | 0.767619 | 0.075828 | 0.110584 | 0.356753 | 0.280935 | ... | 0.174194 | 0.348183 | 0.348195 | 0.824453 | 0.496267 | 3 | 0 | 4 | 4 | 1 |
5 rows × 45 columns
Mixed Logit specification¶
We now make use of the developed formula interface to specify the utilities of the mixed logit model.
We begin by defining the fixed effects parameters, the random effects parameters, and the observed variables. This creates instances of Python objects that can be put together to define the utility functions for the different alternatives.
Once the utilities are defined, we collect them in a Python dictionary mapping alternative names to their corresponding expressions.
from core.dcm_interface import FixedEffect, RandomEffect, ObservedVariable
import torch.distributions as dists
# define fixed effects parameters
B_XF1 = FixedEffect('BETA_XF1')
B_XF2 = FixedEffect('BETA_XF2')
B_XF3 = FixedEffect('BETA_XF3')
# define random effects parameters
B_XR1 = RandomEffect('BETA_XR1')
B_XR2 = RandomEffect('BETA_XR2')
B_XR3 = RandomEffect('BETA_XR3')
B_XR4 = RandomEffect('BETA_XR4')
B_XR5 = RandomEffect('BETA_XR5')
# define observed variables
for attr in df.columns:
exec("%s = ObservedVariable('%s')" % (attr,attr))
# define utility functions
V1 = B_XF1*ALT1_XF1 + B_XF2*ALT1_XF2 + B_XF3*ALT1_XF3 + B_XR1*ALT1_XR1 + B_XR2*ALT1_XR2 + B_XR3*ALT1_XR3 + B_XR4*ALT1_XR4 + B_XR5*ALT1_XR5
V2 = B_XF1*ALT2_XF1 + B_XF2*ALT2_XF2 + B_XF3*ALT2_XF3 + B_XR1*ALT2_XR1 + B_XR2*ALT2_XR2 + B_XR3*ALT2_XR3 + B_XR4*ALT2_XR4 + B_XR5*ALT2_XR5
V3 = B_XF1*ALT3_XF1 + B_XF2*ALT3_XF2 + B_XF3*ALT3_XF3 + B_XR1*ALT3_XR1 + B_XR2*ALT3_XR2 + B_XR3*ALT3_XR3 + B_XR4*ALT3_XR4 + B_XR5*ALT3_XR5
V4 = B_XF1*ALT4_XF1 + B_XF2*ALT4_XF2 + B_XF3*ALT4_XF3 + B_XR1*ALT4_XR1 + B_XR2*ALT4_XR2 + B_XR3*ALT4_XR3 + B_XR4*ALT4_XR4 + B_XR5*ALT4_XR5
V5 = B_XF1*ALT5_XF1 + B_XF2*ALT5_XF2 + B_XF3*ALT5_XF3 + B_XR1*ALT5_XR1 + B_XR2*ALT5_XR2 + B_XR3*ALT5_XR3 + B_XR4*ALT5_XR4 + B_XR5*ALT5_XR5
# associate utility functions with the names of the alternatives
utilities = {"ALT1": V1, "ALT2": V2, "ALT3": V3, "ALT4": V4, "ALT5": V5}
We are now ready to create a Specification object containing the utilities that we have just defined. Note that we must also specify the type of choice model to be used - a mixed logit model (MXL) in this case.
Note that we can inspect the specification by printing the dcm_spec object.
from core.dcm_interface import Specification
# create MXL specification object based on the utilities previously defined
dcm_spec = Specification('MXL', utilities)
print(dcm_spec)
----------------- MXL specification:
Alternatives: ['ALT1', 'ALT2', 'ALT3', 'ALT4', 'ALT5']
Utility functions:
V_ALT1 = BETA_XF1*ALT1_XF1 + BETA_XF2*ALT1_XF2 + BETA_XF3*ALT1_XF3 + BETA_XR1_n*ALT1_XR1 + BETA_XR2_n*ALT1_XR2 + BETA_XR3_n*ALT1_XR3 + BETA_XR4_n*ALT1_XR4 + BETA_XR5_n*ALT1_XR5
V_ALT2 = BETA_XF1*ALT2_XF1 + BETA_XF2*ALT2_XF2 + BETA_XF3*ALT2_XF3 + BETA_XR1_n*ALT2_XR1 + BETA_XR2_n*ALT2_XR2 + BETA_XR3_n*ALT2_XR3 + BETA_XR4_n*ALT2_XR4 + BETA_XR5_n*ALT2_XR5
V_ALT3 = BETA_XF1*ALT3_XF1 + BETA_XF2*ALT3_XF2 + BETA_XF3*ALT3_XF3 + BETA_XR1_n*ALT3_XR1 + BETA_XR2_n*ALT3_XR2 + BETA_XR3_n*ALT3_XR3 + BETA_XR4_n*ALT3_XR4 + BETA_XR5_n*ALT3_XR5
V_ALT4 = BETA_XF1*ALT4_XF1 + BETA_XF2*ALT4_XF2 + BETA_XF3*ALT4_XF3 + BETA_XR1_n*ALT4_XR1 + BETA_XR2_n*ALT4_XR2 + BETA_XR3_n*ALT4_XR3 + BETA_XR4_n*ALT4_XR4 + BETA_XR5_n*ALT4_XR5
V_ALT5 = BETA_XF1*ALT5_XF1 + BETA_XF2*ALT5_XF2 + BETA_XF3*ALT5_XF3 + BETA_XR1_n*ALT5_XR1 + BETA_XR2_n*ALT5_XR2 + BETA_XR3_n*ALT5_XR3 + BETA_XR4_n*ALT5_XR4 + BETA_XR5_n*ALT5_XR5
Num. parameters to be estimated: 8
Fixed effects params: ['BETA_XF1', 'BETA_XF2', 'BETA_XF3']
Random effects params: ['BETA_XR1', 'BETA_XR2', 'BETA_XR3', 'BETA_XR4', 'BETA_XR5']
Once the Specification is defined, we need to define the DCM Dataset object that goes along with it. For this, we instantiate the Dataset class with the Pandas dataframe containing the data in the so-called “wide format”, the name of column in the dataframe containing the observed choices and the dcm_spec that we have previously created.
Note that since this is panel data, we must also specify the name of the column in the dataframe that contains the ID of the respondent (this should be a integer ranging from 0 the num_resp-1).
from core.dcm_interface import Dataset
# create DCM dataset object
dcm_dataset = Dataset(df, 'choice', dcm_spec, resp_id_col='indID')
Preparing dataset...
Model type: MXL
Num. observations: 10000
Num. alternatives: 5
Num. respondents: 2000
Num. menus: 5
Observations IDs: [ 0 1 2 ... 9997 9998 9999]
Alternative IDs: None
Respondent IDs: [ 0 0 0 ... 1999 1999 1999]
Availability columns: None
Attribute names: ['ALT1_XF1', 'ALT1_XF2', 'ALT1_XF3', 'ALT1_XR1', 'ALT1_XR2', 'ALT1_XR3', 'ALT1_XR4', 'ALT1_XR5', 'ALT2_XF1', 'ALT2_XF2', 'ALT2_XF3', 'ALT2_XR1', 'ALT2_XR2', 'ALT2_XR3', 'ALT2_XR4', 'ALT2_XR5', 'ALT3_XF1', 'ALT3_XF2', 'ALT3_XF3', 'ALT3_XR1', 'ALT3_XR2', 'ALT3_XR3', 'ALT3_XR4', 'ALT3_XR5', 'ALT4_XF1', 'ALT4_XF2', 'ALT4_XF3', 'ALT4_XR1', 'ALT4_XR2', 'ALT4_XR3', 'ALT4_XR4', 'ALT4_XR5', 'ALT5_XF1', 'ALT5_XF2', 'ALT5_XF3', 'ALT5_XR1', 'ALT5_XR2', 'ALT5_XR3', 'ALT5_XR4', 'ALT5_XR5']
Fixed effects attribute names: ['ALT1_XF1', 'ALT1_XF2', 'ALT1_XF3', 'ALT2_XF1', 'ALT2_XF2', 'ALT2_XF3', 'ALT3_XF1', 'ALT3_XF2', 'ALT3_XF3', 'ALT4_XF1', 'ALT4_XF2', 'ALT4_XF3', 'ALT5_XF1', 'ALT5_XF2', 'ALT5_XF3']
Fixed effects parameter names: ['BETA_XF1', 'BETA_XF2', 'BETA_XF3', 'BETA_XF1', 'BETA_XF2', 'BETA_XF3', 'BETA_XF1', 'BETA_XF2', 'BETA_XF3', 'BETA_XF1', 'BETA_XF2', 'BETA_XF3', 'BETA_XF1', 'BETA_XF2', 'BETA_XF3']
Random effects attribute names: ['ALT1_XR1', 'ALT1_XR2', 'ALT1_XR3', 'ALT1_XR4', 'ALT1_XR5', 'ALT2_XR1', 'ALT2_XR2', 'ALT2_XR3', 'ALT2_XR4', 'ALT2_XR5', 'ALT3_XR1', 'ALT3_XR2', 'ALT3_XR3', 'ALT3_XR4', 'ALT3_XR5', 'ALT4_XR1', 'ALT4_XR2', 'ALT4_XR3', 'ALT4_XR4', 'ALT4_XR5', 'ALT5_XR1', 'ALT5_XR2', 'ALT5_XR3', 'ALT5_XR4', 'ALT5_XR5']
Random effects parameter names: ['BETA_XR1', 'BETA_XR2', 'BETA_XR3', 'BETA_XR4', 'BETA_XR5', 'BETA_XR1', 'BETA_XR2', 'BETA_XR3', 'BETA_XR4', 'BETA_XR5', 'BETA_XR1', 'BETA_XR2', 'BETA_XR3', 'BETA_XR4', 'BETA_XR5', 'BETA_XR1', 'BETA_XR2', 'BETA_XR3', 'BETA_XR4', 'BETA_XR5', 'BETA_XR1', 'BETA_XR2', 'BETA_XR3', 'BETA_XR4', 'BETA_XR5']
Alternative attributes ndarray.shape: (2000, 5, 40)
Choices ndarray.shape: (2000, 5)
Alternatives availability ndarray.shape: (2000, 5, 5)
Data mask ndarray.shape: (2000, 5)
Context data ndarray.shape: (2000, 0)
Neural nets data ndarray.shape: (2000, 0)
Done!
As with the specification, we can inspect the DCM dataset by printing the dcm_dataset object:
print(dcm_dataset)
----------------- DCM dataset:
Model type: MXL
Num. observations: 10000
Num. alternatives: 5
Num. respondents: 2000
Num. menus: 5
Num. fixed effects: 15
Num. random effects: 25
Attribute names: ['ALT1_XF1', 'ALT1_XF2', 'ALT1_XF3', 'ALT1_XR1', 'ALT1_XR2', 'ALT1_XR3', 'ALT1_XR4', 'ALT1_XR5', 'ALT2_XF1', 'ALT2_XF2', 'ALT2_XF3', 'ALT2_XR1', 'ALT2_XR2', 'ALT2_XR3', 'ALT2_XR4', 'ALT2_XR5', 'ALT3_XF1', 'ALT3_XF2', 'ALT3_XF3', 'ALT3_XR1', 'ALT3_XR2', 'ALT3_XR3', 'ALT3_XR4', 'ALT3_XR5', 'ALT4_XF1', 'ALT4_XF2', 'ALT4_XF3', 'ALT4_XR1', 'ALT4_XR2', 'ALT4_XR3', 'ALT4_XR4', 'ALT4_XR5', 'ALT5_XF1', 'ALT5_XF2', 'ALT5_XF3', 'ALT5_XR1', 'ALT5_XR2', 'ALT5_XR3', 'ALT5_XR4', 'ALT5_XR5']
Bayesian Mixed Logit Model with Automatic Relevance Determination (ARD) in PyTorch¶
We will perform ARD in the MXL model in a similar way as proposed in: Rodrigues, F., Ortelli, N., Bierlaire, M. and Pereira, F.C., 2020. Bayesian automatic relevance determination for utility function specification in discrete choice models. IEEE Transactions on Intelligent Transportation Systems.
We begin by modifying the generative process of the core MXL model as follows (changes to the MXL model are highlighted in red): \(\require{color}\)
Draw variance for prior over fixed taste parameters \(\boldsymbol\tau_\alpha \sim \mbox{InverseGamma}(\alpha_0, \beta_0)\)
Draw fixed taste parameters \(\boldsymbol\alpha \sim \mathcal{N}(\boldsymbol\lambda_0, \color{red} \mbox{diag}(\boldsymbol\tau_\zeta) \color{black})\)
Draw variance for prior over random taste parameters \(\boldsymbol\tau_\zeta \sim \mbox{InverseGamma}(\alpha_0, \beta_0)\)
Draw mean vector \(\boldsymbol\zeta \sim \mathcal{N}(\boldsymbol\mu_0, \color{red} \mbox{diag}(\boldsymbol\tau_\zeta) \color{black})\)
Draw scales vector \(\boldsymbol\theta \sim \mbox{half-Cauchy}(\boldsymbol\sigma_0)\)
Draw correlation matrix \(\boldsymbol\Psi \sim \mbox{LKJ}(\nu)\)
For each decision-maker \(n \in \{1,\dots,N\}\)
Draw random taste parameters \(\boldsymbol\beta_n \sim \mathcal{N}(\boldsymbol\zeta,\boldsymbol\Omega)\)
For each choice occasion \(t \in \{1,\dots,T_n\}\)
Draw observed choice \(y_{nt} \sim \mbox{MNL}(\boldsymbol\alpha, \boldsymbol\beta_n, \textbf{X}_{nt})\)
where \(\boldsymbol\Omega = \mbox{diag}(\boldsymbol\theta) \times \boldsymbol\Psi \times \mbox{diag}(\boldsymbol\theta)\).
This model is already implemented in the class TorchMXL_ARD. At the end of this notebook, we provide an explanation of how this extension was implemented.
We can instantiate this model from the TorchMXL_ARD using the following code. We can the run variational inference to approximate the posterior distribution of the latent variables in the model. Note that since in this case we know the true parameters that were used to generate the simualated choice data, we can pass them to the “infer” method in order to obtain additional information during the ELBO maximization (useful for tracking the progress of VI and for other debugging purposes).
%%time
from core.torch_mxl_ard import TorchMXL_ARD
# instantiate MXL model
mxl = TorchMXL_ARD(dcm_dataset, batch_size=num_resp, use_inference_net=False, use_cuda=True)
# run Bayesian inference (variational inference)
results = mxl.infer(num_epochs=20000, true_alpha=true_alpha, true_beta=true_beta)
[Epoch 0] ELBO: 30651; Loglik: -21061; Acc.: 0.169; Alpha RMSE: 0.645; Beta RMSE: 0.765
[Epoch 100] ELBO: 36925; Loglik: -16834; Acc.: 0.277; Alpha RMSE: 0.265; Beta RMSE: 0.788
[Epoch 200] ELBO: 28090; Loglik: -15822; Acc.: 0.326; Alpha RMSE: 0.091; Beta RMSE: 0.773
[Epoch 300] ELBO: 18901; Loglik: -14839; Acc.: 0.376; Alpha RMSE: 0.048; Beta RMSE: 0.725
[Epoch 400] ELBO: 19277; Loglik: -14460; Acc.: 0.388; Alpha RMSE: 0.086; Beta RMSE: 0.699
[Epoch 500] ELBO: 18957; Loglik: -13972; Acc.: 0.414; Alpha RMSE: 0.082; Beta RMSE: 0.623
[Epoch 600] ELBO: 17863; Loglik: -13922; Acc.: 0.417; Alpha RMSE: 0.068; Beta RMSE: 0.594
[Epoch 700] ELBO: 16851; Loglik: -13622; Acc.: 0.436; Alpha RMSE: 0.073; Beta RMSE: 0.549
[Epoch 800] ELBO: 16725; Loglik: -13468; Acc.: 0.443; Alpha RMSE: 0.124; Beta RMSE: 0.502
[Epoch 900] ELBO: 16681; Loglik: -13089; Acc.: 0.458; Alpha RMSE: 0.101; Beta RMSE: 0.446
[Epoch 1000] ELBO: 16433; Loglik: -13137; Acc.: 0.463; Alpha RMSE: 0.117; Beta RMSE: 0.407
[Epoch 1100] ELBO: 16039; Loglik: -12896; Acc.: 0.475; Alpha RMSE: 0.132; Beta RMSE: 0.357
[Epoch 1200] ELBO: 16181; Loglik: -12948; Acc.: 0.467; Alpha RMSE: 0.134; Beta RMSE: 0.318
[Epoch 1300] ELBO: 16435; Loglik: -13016; Acc.: 0.466; Alpha RMSE: 0.134; Beta RMSE: 0.291
[Epoch 1400] ELBO: 15930; Loglik: -12756; Acc.: 0.473; Alpha RMSE: 0.153; Beta RMSE: 0.253
[Epoch 1500] ELBO: 15771; Loglik: -12708; Acc.: 0.480; Alpha RMSE: 0.150; Beta RMSE: 0.223
[Epoch 1600] ELBO: 15783; Loglik: -12719; Acc.: 0.483; Alpha RMSE: 0.152; Beta RMSE: 0.201
[Epoch 1700] ELBO: 15770; Loglik: -12713; Acc.: 0.475; Alpha RMSE: 0.150; Beta RMSE: 0.156
[Epoch 1800] ELBO: 15704; Loglik: -12737; Acc.: 0.477; Alpha RMSE: 0.144; Beta RMSE: 0.127
[Epoch 1900] ELBO: 15666; Loglik: -12626; Acc.: 0.485; Alpha RMSE: 0.159; Beta RMSE: 0.090
[Epoch 2000] ELBO: 15738; Loglik: -12645; Acc.: 0.488; Alpha RMSE: 0.164; Beta RMSE: 0.082
[Epoch 2100] ELBO: 15656; Loglik: -12618; Acc.: 0.486; Alpha RMSE: 0.154; Beta RMSE: 0.084
[Epoch 2200] ELBO: 15617; Loglik: -12564; Acc.: 0.482; Alpha RMSE: 0.165; Beta RMSE: 0.081
[Epoch 2300] ELBO: 15517; Loglik: -12605; Acc.: 0.484; Alpha RMSE: 0.161; Beta RMSE: 0.083
[Epoch 2400] ELBO: 15389; Loglik: -12629; Acc.: 0.488; Alpha RMSE: 0.149; Beta RMSE: 0.084
[Epoch 2500] ELBO: 15326; Loglik: -12661; Acc.: 0.488; Alpha RMSE: 0.161; Beta RMSE: 0.092
[Epoch 2600] ELBO: 15370; Loglik: -12631; Acc.: 0.484; Alpha RMSE: 0.139; Beta RMSE: 0.102
[Epoch 2700] ELBO: 15366; Loglik: -12793; Acc.: 0.484; Alpha RMSE: 0.135; Beta RMSE: 0.115
[Epoch 2800] ELBO: 15255; Loglik: -12647; Acc.: 0.483; Alpha RMSE: 0.152; Beta RMSE: 0.117
[Epoch 2900] ELBO: 15538; Loglik: -12722; Acc.: 0.478; Alpha RMSE: 0.148; Beta RMSE: 0.100
[Epoch 3000] ELBO: 15273; Loglik: -12792; Acc.: 0.478; Alpha RMSE: 0.149; Beta RMSE: 0.122
[Epoch 3100] ELBO: 15169; Loglik: -12714; Acc.: 0.484; Alpha RMSE: 0.142; Beta RMSE: 0.130
[Epoch 3200] ELBO: 15295; Loglik: -12820; Acc.: 0.481; Alpha RMSE: 0.133; Beta RMSE: 0.124
[Epoch 3300] ELBO: 15447; Loglik: -12823; Acc.: 0.476; Alpha RMSE: 0.137; Beta RMSE: 0.129
[Epoch 3400] ELBO: 15348; Loglik: -12769; Acc.: 0.475; Alpha RMSE: 0.128; Beta RMSE: 0.123
[Epoch 3500] ELBO: 15313; Loglik: -12796; Acc.: 0.471; Alpha RMSE: 0.121; Beta RMSE: 0.132
[Epoch 3600] ELBO: 15236; Loglik: -12797; Acc.: 0.473; Alpha RMSE: 0.130; Beta RMSE: 0.116
[Epoch 3700] ELBO: 15085; Loglik: -12827; Acc.: 0.473; Alpha RMSE: 0.131; Beta RMSE: 0.120
[Epoch 3800] ELBO: 15123; Loglik: -12819; Acc.: 0.475; Alpha RMSE: 0.131; Beta RMSE: 0.109
[Epoch 3900] ELBO: 15199; Loglik: -12880; Acc.: 0.474; Alpha RMSE: 0.126; Beta RMSE: 0.120
[Epoch 4000] ELBO: 15179; Loglik: -12964; Acc.: 0.464; Alpha RMSE: 0.125; Beta RMSE: 0.104
[Epoch 4100] ELBO: 15123; Loglik: -12944; Acc.: 0.460; Alpha RMSE: 0.130; Beta RMSE: 0.110
[Epoch 4200] ELBO: 15139; Loglik: -12957; Acc.: 0.469; Alpha RMSE: 0.115; Beta RMSE: 0.099
[Epoch 4300] ELBO: 14954; Loglik: -12867; Acc.: 0.468; Alpha RMSE: 0.113; Beta RMSE: 0.115
[Epoch 4400] ELBO: 15007; Loglik: -12925; Acc.: 0.467; Alpha RMSE: 0.120; Beta RMSE: 0.094
[Epoch 4500] ELBO: 15044; Loglik: -12987; Acc.: 0.462; Alpha RMSE: 0.110; Beta RMSE: 0.106
[Epoch 4600] ELBO: 15055; Loglik: -13000; Acc.: 0.461; Alpha RMSE: 0.111; Beta RMSE: 0.091
[Epoch 4700] ELBO: 14921; Loglik: -12953; Acc.: 0.468; Alpha RMSE: 0.099; Beta RMSE: 0.087
[Epoch 4800] ELBO: 14981; Loglik: -12959; Acc.: 0.463; Alpha RMSE: 0.112; Beta RMSE: 0.084
[Epoch 4900] ELBO: 15000; Loglik: -12967; Acc.: 0.460; Alpha RMSE: 0.104; Beta RMSE: 0.091
[Epoch 5000] ELBO: 14971; Loglik: -13115; Acc.: 0.458; Alpha RMSE: 0.095; Beta RMSE: 0.074
[Epoch 5100] ELBO: 14842; Loglik: -12976; Acc.: 0.461; Alpha RMSE: 0.092; Beta RMSE: 0.081
[Epoch 5200] ELBO: 14916; Loglik: -13087; Acc.: 0.461; Alpha RMSE: 0.095; Beta RMSE: 0.070
[Epoch 5300] ELBO: 14887; Loglik: -13059; Acc.: 0.459; Alpha RMSE: 0.109; Beta RMSE: 0.076
[Epoch 5400] ELBO: 14920; Loglik: -13117; Acc.: 0.455; Alpha RMSE: 0.094; Beta RMSE: 0.062
[Epoch 5500] ELBO: 14921; Loglik: -13093; Acc.: 0.452; Alpha RMSE: 0.098; Beta RMSE: 0.062
[Epoch 5600] ELBO: 14876; Loglik: -13173; Acc.: 0.451; Alpha RMSE: 0.101; Beta RMSE: 0.069
[Epoch 5700] ELBO: 14884; Loglik: -13155; Acc.: 0.451; Alpha RMSE: 0.079; Beta RMSE: 0.074
[Epoch 5800] ELBO: 14883; Loglik: -13131; Acc.: 0.454; Alpha RMSE: 0.101; Beta RMSE: 0.067
[Epoch 5900] ELBO: 14907; Loglik: -13239; Acc.: 0.454; Alpha RMSE: 0.083; Beta RMSE: 0.063
[Epoch 6000] ELBO: 14886; Loglik: -13209; Acc.: 0.449; Alpha RMSE: 0.088; Beta RMSE: 0.064
[Epoch 6100] ELBO: 14897; Loglik: -13202; Acc.: 0.459; Alpha RMSE: 0.085; Beta RMSE: 0.057
[Epoch 6200] ELBO: 14775; Loglik: -13161; Acc.: 0.451; Alpha RMSE: 0.075; Beta RMSE: 0.061
[Epoch 6300] ELBO: 14811; Loglik: -13200; Acc.: 0.453; Alpha RMSE: 0.089; Beta RMSE: 0.058
[Epoch 6400] ELBO: 14890; Loglik: -13291; Acc.: 0.443; Alpha RMSE: 0.081; Beta RMSE: 0.050
[Epoch 6500] ELBO: 14804; Loglik: -13226; Acc.: 0.454; Alpha RMSE: 0.078; Beta RMSE: 0.060
[Epoch 6600] ELBO: 14912; Loglik: -13289; Acc.: 0.447; Alpha RMSE: 0.093; Beta RMSE: 0.042
[Epoch 6700] ELBO: 14749; Loglik: -13182; Acc.: 0.451; Alpha RMSE: 0.081; Beta RMSE: 0.040
[Epoch 6800] ELBO: 14824; Loglik: -13285; Acc.: 0.444; Alpha RMSE: 0.080; Beta RMSE: 0.051
[Epoch 6900] ELBO: 14835; Loglik: -13323; Acc.: 0.444; Alpha RMSE: 0.083; Beta RMSE: 0.041
[Epoch 7000] ELBO: 14910; Loglik: -13372; Acc.: 0.443; Alpha RMSE: 0.072; Beta RMSE: 0.038
[Epoch 7100] ELBO: 14906; Loglik: -13361; Acc.: 0.442; Alpha RMSE: 0.085; Beta RMSE: 0.042
[Epoch 7200] ELBO: 14867; Loglik: -13382; Acc.: 0.438; Alpha RMSE: 0.078; Beta RMSE: 0.031
[Epoch 7300] ELBO: 14705; Loglik: -13209; Acc.: 0.452; Alpha RMSE: 0.074; Beta RMSE: 0.043
[Epoch 7400] ELBO: 14862; Loglik: -13389; Acc.: 0.439; Alpha RMSE: 0.081; Beta RMSE: 0.033
[Epoch 7500] ELBO: 14812; Loglik: -13349; Acc.: 0.447; Alpha RMSE: 0.090; Beta RMSE: 0.036
[Epoch 7600] ELBO: 14754; Loglik: -13255; Acc.: 0.448; Alpha RMSE: 0.074; Beta RMSE: 0.034
[Epoch 7700] ELBO: 14827; Loglik: -13398; Acc.: 0.438; Alpha RMSE: 0.068; Beta RMSE: 0.039
[Epoch 7800] ELBO: 14874; Loglik: -13431; Acc.: 0.438; Alpha RMSE: 0.070; Beta RMSE: 0.032
[Epoch 7900] ELBO: 14815; Loglik: -13383; Acc.: 0.440; Alpha RMSE: 0.083; Beta RMSE: 0.033
[Epoch 8000] ELBO: 14843; Loglik: -13410; Acc.: 0.437; Alpha RMSE: 0.077; Beta RMSE: 0.033
[Epoch 8100] ELBO: 14827; Loglik: -13422; Acc.: 0.438; Alpha RMSE: 0.071; Beta RMSE: 0.030
[Epoch 8200] ELBO: 14830; Loglik: -13422; Acc.: 0.436; Alpha RMSE: 0.090; Beta RMSE: 0.028
[Epoch 8300] ELBO: 14794; Loglik: -13385; Acc.: 0.440; Alpha RMSE: 0.069; Beta RMSE: 0.031
[Epoch 8400] ELBO: 14797; Loglik: -13404; Acc.: 0.443; Alpha RMSE: 0.063; Beta RMSE: 0.024
[Epoch 8500] ELBO: 14901; Loglik: -13502; Acc.: 0.432; Alpha RMSE: 0.054; Beta RMSE: 0.032
[Epoch 8600] ELBO: 14889; Loglik: -13485; Acc.: 0.439; Alpha RMSE: 0.070; Beta RMSE: 0.023
[Epoch 8700] ELBO: 14873; Loglik: -13496; Acc.: 0.431; Alpha RMSE: 0.082; Beta RMSE: 0.023
[Epoch 8800] ELBO: 14794; Loglik: -13445; Acc.: 0.433; Alpha RMSE: 0.078; Beta RMSE: 0.021
[Epoch 8900] ELBO: 14805; Loglik: -13447; Acc.: 0.437; Alpha RMSE: 0.072; Beta RMSE: 0.025
[Epoch 9000] ELBO: 14775; Loglik: -13415; Acc.: 0.432; Alpha RMSE: 0.056; Beta RMSE: 0.022
[Epoch 9100] ELBO: 14774; Loglik: -13435; Acc.: 0.438; Alpha RMSE: 0.064; Beta RMSE: 0.021
[Epoch 9200] ELBO: 14752; Loglik: -13421; Acc.: 0.444; Alpha RMSE: 0.073; Beta RMSE: 0.023
[Epoch 9300] ELBO: 14797; Loglik: -13464; Acc.: 0.433; Alpha RMSE: 0.057; Beta RMSE: 0.019
[Epoch 9400] ELBO: 14849; Loglik: -13523; Acc.: 0.437; Alpha RMSE: 0.077; Beta RMSE: 0.020
[Epoch 9500] ELBO: 14794; Loglik: -13472; Acc.: 0.438; Alpha RMSE: 0.069; Beta RMSE: 0.020
[Epoch 9600] ELBO: 14820; Loglik: -13505; Acc.: 0.431; Alpha RMSE: 0.063; Beta RMSE: 0.015
[Epoch 9700] ELBO: 14803; Loglik: -13479; Acc.: 0.434; Alpha RMSE: 0.059; Beta RMSE: 0.013
[Epoch 9800] ELBO: 14823; Loglik: -13506; Acc.: 0.433; Alpha RMSE: 0.065; Beta RMSE: 0.018
[Epoch 9900] ELBO: 14842; Loglik: -13523; Acc.: 0.436; Alpha RMSE: 0.078; Beta RMSE: 0.014
[Epoch 10000] ELBO: 14758; Loglik: -13449; Acc.: 0.435; Alpha RMSE: 0.062; Beta RMSE: 0.021
[Epoch 10100] ELBO: 14728; Loglik: -13420; Acc.: 0.438; Alpha RMSE: 0.046; Beta RMSE: 0.012
[Epoch 10200] ELBO: 14710; Loglik: -13413; Acc.: 0.439; Alpha RMSE: 0.049; Beta RMSE: 0.021
[Epoch 10300] ELBO: 14775; Loglik: -13471; Acc.: 0.439; Alpha RMSE: 0.074; Beta RMSE: 0.012
[Epoch 10400] ELBO: 14796; Loglik: -13493; Acc.: 0.439; Alpha RMSE: 0.063; Beta RMSE: 0.020
[Epoch 10500] ELBO: 14744; Loglik: -13461; Acc.: 0.437; Alpha RMSE: 0.069; Beta RMSE: 0.019
[Epoch 10600] ELBO: 14775; Loglik: -13475; Acc.: 0.438; Alpha RMSE: 0.077; Beta RMSE: 0.018
[Epoch 10700] ELBO: 14806; Loglik: -13515; Acc.: 0.431; Alpha RMSE: 0.066; Beta RMSE: 0.022
[Epoch 10800] ELBO: 14827; Loglik: -13534; Acc.: 0.437; Alpha RMSE: 0.072; Beta RMSE: 0.017
[Epoch 10900] ELBO: 14763; Loglik: -13478; Acc.: 0.446; Alpha RMSE: 0.073; Beta RMSE: 0.014
[Epoch 11000] ELBO: 14781; Loglik: -13489; Acc.: 0.432; Alpha RMSE: 0.061; Beta RMSE: 0.014
[Epoch 11100] ELBO: 14752; Loglik: -13467; Acc.: 0.437; Alpha RMSE: 0.072; Beta RMSE: 0.013
[Epoch 11200] ELBO: 14878; Loglik: -13590; Acc.: 0.424; Alpha RMSE: 0.060; Beta RMSE: 0.015
[Epoch 11300] ELBO: 14820; Loglik: -13535; Acc.: 0.432; Alpha RMSE: 0.063; Beta RMSE: 0.021
[Epoch 11400] ELBO: 14842; Loglik: -13567; Acc.: 0.428; Alpha RMSE: 0.056; Beta RMSE: 0.016
[Epoch 11500] ELBO: 14856; Loglik: -13592; Acc.: 0.428; Alpha RMSE: 0.076; Beta RMSE: 0.015
[Epoch 11600] ELBO: 14789; Loglik: -13511; Acc.: 0.441; Alpha RMSE: 0.065; Beta RMSE: 0.018
[Epoch 11700] ELBO: 14773; Loglik: -13499; Acc.: 0.434; Alpha RMSE: 0.066; Beta RMSE: 0.023
[Epoch 11800] ELBO: 14789; Loglik: -13533; Acc.: 0.433; Alpha RMSE: 0.061; Beta RMSE: 0.013
[Epoch 11900] ELBO: 14751; Loglik: -13487; Acc.: 0.442; Alpha RMSE: 0.047; Beta RMSE: 0.019
[Epoch 12000] ELBO: 14792; Loglik: -13528; Acc.: 0.435; Alpha RMSE: 0.072; Beta RMSE: 0.019
[Epoch 12100] ELBO: 14791; Loglik: -13543; Acc.: 0.435; Alpha RMSE: 0.065; Beta RMSE: 0.016
[Epoch 12200] ELBO: 14760; Loglik: -13499; Acc.: 0.434; Alpha RMSE: 0.069; Beta RMSE: 0.015
[Epoch 12300] ELBO: 14765; Loglik: -13509; Acc.: 0.439; Alpha RMSE: 0.074; Beta RMSE: 0.016
[Epoch 12400] ELBO: 14833; Loglik: -13574; Acc.: 0.431; Alpha RMSE: 0.056; Beta RMSE: 0.015
[Epoch 12500] ELBO: 14883; Loglik: -13622; Acc.: 0.430; Alpha RMSE: 0.055; Beta RMSE: 0.024
[Epoch 12600] ELBO: 14780; Loglik: -13521; Acc.: 0.435; Alpha RMSE: 0.068; Beta RMSE: 0.014
[Epoch 12700] ELBO: 14775; Loglik: -13518; Acc.: 0.433; Alpha RMSE: 0.067; Beta RMSE: 0.015
[Epoch 12800] ELBO: 14854; Loglik: -13600; Acc.: 0.434; Alpha RMSE: 0.070; Beta RMSE: 0.021
[Epoch 12900] ELBO: 14818; Loglik: -13567; Acc.: 0.433; Alpha RMSE: 0.058; Beta RMSE: 0.018
[Epoch 13000] ELBO: 14772; Loglik: -13503; Acc.: 0.433; Alpha RMSE: 0.068; Beta RMSE: 0.020
[Epoch 13100] ELBO: 14829; Loglik: -13588; Acc.: 0.426; Alpha RMSE: 0.066; Beta RMSE: 0.015
[Epoch 13200] ELBO: 14753; Loglik: -13493; Acc.: 0.434; Alpha RMSE: 0.075; Beta RMSE: 0.016
[Epoch 13300] ELBO: 14876; Loglik: -13633; Acc.: 0.424; Alpha RMSE: 0.074; Beta RMSE: 0.014
[Epoch 13400] ELBO: 14802; Loglik: -13541; Acc.: 0.437; Alpha RMSE: 0.054; Beta RMSE: 0.015
[Epoch 13500] ELBO: 14763; Loglik: -13511; Acc.: 0.432; Alpha RMSE: 0.067; Beta RMSE: 0.015
[Epoch 13600] ELBO: 14861; Loglik: -13609; Acc.: 0.430; Alpha RMSE: 0.063; Beta RMSE: 0.012
[Epoch 13700] ELBO: 14793; Loglik: -13551; Acc.: 0.438; Alpha RMSE: 0.059; Beta RMSE: 0.014
[Epoch 13800] ELBO: 14752; Loglik: -13513; Acc.: 0.435; Alpha RMSE: 0.059; Beta RMSE: 0.010
[Epoch 13900] ELBO: 14813; Loglik: -13567; Acc.: 0.432; Alpha RMSE: 0.072; Beta RMSE: 0.020
[Epoch 14000] ELBO: 14793; Loglik: -13551; Acc.: 0.427; Alpha RMSE: 0.063; Beta RMSE: 0.014
[Epoch 14100] ELBO: 14883; Loglik: -13637; Acc.: 0.429; Alpha RMSE: 0.079; Beta RMSE: 0.014
[Epoch 14200] ELBO: 14779; Loglik: -13530; Acc.: 0.433; Alpha RMSE: 0.056; Beta RMSE: 0.019
[Epoch 14300] ELBO: 14774; Loglik: -13518; Acc.: 0.428; Alpha RMSE: 0.061; Beta RMSE: 0.023
[Epoch 14400] ELBO: 14738; Loglik: -13488; Acc.: 0.432; Alpha RMSE: 0.051; Beta RMSE: 0.016
[Epoch 14500] ELBO: 14791; Loglik: -13541; Acc.: 0.431; Alpha RMSE: 0.054; Beta RMSE: 0.014
[Epoch 14600] ELBO: 14829; Loglik: -13594; Acc.: 0.427; Alpha RMSE: 0.077; Beta RMSE: 0.011
[Epoch 14700] ELBO: 14836; Loglik: -13612; Acc.: 0.428; Alpha RMSE: 0.071; Beta RMSE: 0.015
[Epoch 14800] ELBO: 14809; Loglik: -13572; Acc.: 0.431; Alpha RMSE: 0.061; Beta RMSE: 0.016
[Epoch 14900] ELBO: 14761; Loglik: -13528; Acc.: 0.428; Alpha RMSE: 0.057; Beta RMSE: 0.012
[Epoch 15000] ELBO: 14771; Loglik: -13530; Acc.: 0.428; Alpha RMSE: 0.052; Beta RMSE: 0.017
[Epoch 15100] ELBO: 14793; Loglik: -13553; Acc.: 0.431; Alpha RMSE: 0.061; Beta RMSE: 0.013
[Epoch 15200] ELBO: 14826; Loglik: -13580; Acc.: 0.429; Alpha RMSE: 0.070; Beta RMSE: 0.019
[Epoch 15300] ELBO: 14753; Loglik: -13506; Acc.: 0.431; Alpha RMSE: 0.052; Beta RMSE: 0.017
[Epoch 15400] ELBO: 14805; Loglik: -13561; Acc.: 0.434; Alpha RMSE: 0.049; Beta RMSE: 0.013
[Epoch 15500] ELBO: 14804; Loglik: -13562; Acc.: 0.428; Alpha RMSE: 0.054; Beta RMSE: 0.012
[Epoch 15600] ELBO: 14847; Loglik: -13617; Acc.: 0.428; Alpha RMSE: 0.074; Beta RMSE: 0.014
[Epoch 15700] ELBO: 14851; Loglik: -13623; Acc.: 0.433; Alpha RMSE: 0.065; Beta RMSE: 0.011
[Epoch 15800] ELBO: 14829; Loglik: -13588; Acc.: 0.427; Alpha RMSE: 0.065; Beta RMSE: 0.016
[Epoch 15900] ELBO: 14788; Loglik: -13553; Acc.: 0.429; Alpha RMSE: 0.062; Beta RMSE: 0.012
[Epoch 16000] ELBO: 14825; Loglik: -13595; Acc.: 0.427; Alpha RMSE: 0.052; Beta RMSE: 0.012
[Epoch 16100] ELBO: 14703; Loglik: -13474; Acc.: 0.438; Alpha RMSE: 0.051; Beta RMSE: 0.016
[Epoch 16200] ELBO: 14768; Loglik: -13524; Acc.: 0.433; Alpha RMSE: 0.055; Beta RMSE: 0.015
[Epoch 16300] ELBO: 14757; Loglik: -13520; Acc.: 0.431; Alpha RMSE: 0.070; Beta RMSE: 0.012
[Epoch 16400] ELBO: 14798; Loglik: -13561; Acc.: 0.434; Alpha RMSE: 0.063; Beta RMSE: 0.013
[Epoch 16500] ELBO: 14751; Loglik: -13510; Acc.: 0.429; Alpha RMSE: 0.057; Beta RMSE: 0.009
[Epoch 16600] ELBO: 14805; Loglik: -13566; Acc.: 0.434; Alpha RMSE: 0.052; Beta RMSE: 0.011
[Epoch 16700] ELBO: 14821; Loglik: -13582; Acc.: 0.426; Alpha RMSE: 0.066; Beta RMSE: 0.011
[Epoch 16800] ELBO: 14826; Loglik: -13601; Acc.: 0.427; Alpha RMSE: 0.052; Beta RMSE: 0.013
[Epoch 16900] ELBO: 14777; Loglik: -13553; Acc.: 0.429; Alpha RMSE: 0.057; Beta RMSE: 0.008
[Epoch 17000] ELBO: 14770; Loglik: -13536; Acc.: 0.437; Alpha RMSE: 0.065; Beta RMSE: 0.016
[Epoch 17100] ELBO: 14800; Loglik: -13570; Acc.: 0.431; Alpha RMSE: 0.057; Beta RMSE: 0.011
[Epoch 17200] ELBO: 14822; Loglik: -13589; Acc.: 0.435; Alpha RMSE: 0.065; Beta RMSE: 0.012
[Epoch 17300] ELBO: 14799; Loglik: -13565; Acc.: 0.430; Alpha RMSE: 0.063; Beta RMSE: 0.016
[Epoch 17400] ELBO: 14832; Loglik: -13598; Acc.: 0.429; Alpha RMSE: 0.053; Beta RMSE: 0.014
[Epoch 17500] ELBO: 14785; Loglik: -13550; Acc.: 0.432; Alpha RMSE: 0.074; Beta RMSE: 0.017
[Epoch 17600] ELBO: 14815; Loglik: -13575; Acc.: 0.428; Alpha RMSE: 0.050; Beta RMSE: 0.014
[Epoch 17700] ELBO: 14831; Loglik: -13591; Acc.: 0.436; Alpha RMSE: 0.059; Beta RMSE: 0.019
[Epoch 17800] ELBO: 14801; Loglik: -13564; Acc.: 0.433; Alpha RMSE: 0.064; Beta RMSE: 0.015
[Epoch 17900] ELBO: 14805; Loglik: -13584; Acc.: 0.436; Alpha RMSE: 0.075; Beta RMSE: 0.013
[Epoch 18000] ELBO: 14697; Loglik: -13468; Acc.: 0.439; Alpha RMSE: 0.051; Beta RMSE: 0.012
[Epoch 18100] ELBO: 14773; Loglik: -13553; Acc.: 0.431; Alpha RMSE: 0.067; Beta RMSE: 0.010
[Epoch 18200] ELBO: 14837; Loglik: -13610; Acc.: 0.422; Alpha RMSE: 0.048; Beta RMSE: 0.014
[Epoch 18300] ELBO: 14815; Loglik: -13585; Acc.: 0.437; Alpha RMSE: 0.072; Beta RMSE: 0.015
[Epoch 18400] ELBO: 14792; Loglik: -13555; Acc.: 0.427; Alpha RMSE: 0.060; Beta RMSE: 0.017
[Epoch 18500] ELBO: 14788; Loglik: -13562; Acc.: 0.434; Alpha RMSE: 0.067; Beta RMSE: 0.020
[Epoch 18600] ELBO: 14728; Loglik: -13493; Acc.: 0.434; Alpha RMSE: 0.059; Beta RMSE: 0.015
[Epoch 18700] ELBO: 14827; Loglik: -13605; Acc.: 0.429; Alpha RMSE: 0.053; Beta RMSE: 0.013
[Epoch 18800] ELBO: 14785; Loglik: -13549; Acc.: 0.432; Alpha RMSE: 0.068; Beta RMSE: 0.012
[Epoch 18900] ELBO: 14805; Loglik: -13567; Acc.: 0.428; Alpha RMSE: 0.056; Beta RMSE: 0.007
[Epoch 19000] ELBO: 14793; Loglik: -13559; Acc.: 0.433; Alpha RMSE: 0.060; Beta RMSE: 0.008
[Epoch 19100] ELBO: 14833; Loglik: -13595; Acc.: 0.431; Alpha RMSE: 0.069; Beta RMSE: 0.014
[Epoch 19200] ELBO: 14822; Loglik: -13593; Acc.: 0.429; Alpha RMSE: 0.063; Beta RMSE: 0.014
[Epoch 19300] ELBO: 14805; Loglik: -13572; Acc.: 0.431; Alpha RMSE: 0.048; Beta RMSE: 0.015
[Epoch 19400] ELBO: 14766; Loglik: -13549; Acc.: 0.431; Alpha RMSE: 0.069; Beta RMSE: 0.017
[Epoch 19500] ELBO: 14783; Loglik: -13545; Acc.: 0.433; Alpha RMSE: 0.060; Beta RMSE: 0.009
[Epoch 19600] ELBO: 14766; Loglik: -13537; Acc.: 0.434; Alpha RMSE: 0.073; Beta RMSE: 0.013
[Epoch 19700] ELBO: 14747; Loglik: -13520; Acc.: 0.433; Alpha RMSE: 0.042; Beta RMSE: 0.012
[Epoch 19800] ELBO: 14812; Loglik: -13583; Acc.: 0.430; Alpha RMSE: 0.057; Beta RMSE: 0.013
[Epoch 19900] ELBO: 14804; Loglik: -13581; Acc.: 0.427; Alpha RMSE: 0.089; Beta RMSE: 0.013
Elapsed time: 304.10623359680176
True alpha: [-0.8 0.8 0. ]
Est. alpha: [-8.2980311e-01 8.8909477e-01 5.0747342e-04]
BETA_XF1: -0.830
BETA_XF2: 0.889
BETA_XF3: 0.001
True zeta: [-0.8 0.8 1. -0.8 0. ]
Est. zeta: [-0.817338 0.7997669 0.9919458 -0.81087536 -0.00533627]
BETA_XR1: -0.817
BETA_XR2: 0.800
BETA_XR3: 0.992
BETA_XR4: -0.811
BETA_XR5: -0.005
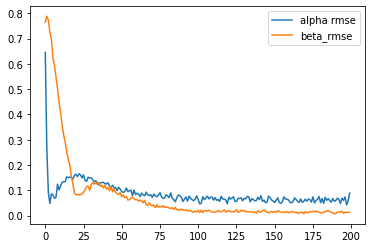
CPU times: user 53min, sys: 2.58 s, total: 53min 3s
Wall time: 5min 7s
Lets now have a look at the inferred prior variances. Irrelevant components in the utility will have the expected values of the prior variances of their corresponding parameters shrunk to values close to zero. Starting with the expected values of the prior variances for the fixed effects:
1/(mxl.softplus(mxl.tau_alpha_alpha) / mxl.softplus(mxl.tau_alpha_beta))
tensor([1.0889, 1.0683, 0.0384], device='cuda:0', grad_fn=<MulBackward0>)
We can see that the last preference parameter has a value close to zero, which indicates that the inclusion of that term in the utility function is irrelevant. This is the desired behaviour given the way that the data was generated.
Lets now have a look at the expected values of the prior variances for the random effects:
1/(mxl.softplus(mxl.tau_zeta_alpha) / mxl.softplus(mxl.tau_zeta_beta))
tensor([1.0584, 1.0380, 1.3097, 1.0221, 0.0225], device='cuda:0',
grad_fn=<MulBackward0>)
As before, can see that the last preference parameter has a value close to zero, which indicates that the inclusion of that term in the utility function is irrelevant. This is the desired behaviour given the way that the data was generated.
The “results” dictionary containts a summary of the results of variational inference, including means of the posterior approximations for the different parameters in the model:
results
{'Estimation time': 304.10623359680176,
'Est. alpha': array([-8.2980311e-01, 8.8909477e-01, 5.0747342e-04], dtype=float32),
'Est. zeta': array([-0.817338 , 0.7997669 , 0.9919458 , -0.81087536, -0.00533627],
dtype=float32),
'Est. beta_n': array([[-1.238147 , 0.35534793, 0.56024605, -0.9264143 , -0.441626 ],
[-0.5745481 , 1.3353534 , 1.3706696 , -0.3979419 , 0.5681873 ],
[-1.7197372 , -0.48435944, 0.08551951, -1.7841139 , -0.9252483 ],
...,
[ 0.24848358, 2.0946019 , 2.317096 , 0.45183292, 1.5201857 ],
[-0.81808007, 0.84409773, 1.0342833 , -0.71131504, -0.04303875],
[-0.6825954 , 1.1738247 , 1.2203168 , -0.5835304 , 0.14842193]],
dtype=float32),
'ELBO': 14744.6484375,
'Loglikelihood': -13506.572265625,
'Accuracy': 0.43320000171661377}
This interface is currently being improved to include additional output information, but additional information can be obtained from the attributes of the “mxl” object for now.
Implementation details¶
Modifying the core MXL model implementation with this ARD extension is relatively straightforward. The generative process described above is different from the core MXL model, so we begin by introducing the two new priors (\(\boldsymbol\tau_\alpha\) and \(\boldsymbol\tau_\zeta\)) in the elbo() function:
tau_alpha_prior = td.Gamma(.01*torch.ones(self.num_fixed_params, device=self.device),
.01*torch.ones(self.num_fixed_params, device=self.device))
tau_zeta_prior = td.Gamma(.01*torch.ones(self.num_mixed_params, device=self.device),
.01*torch.ones(self.num_mixed_params, device=self.device))
Note that we use the Gamma distribution, but we will later take the inverse of its samples.
Now we must also change the priors over \(\boldsymbol\alpha\) and \(\boldsymbol\zeta\) to use the newly defined priors over the variances:
alpha_prior = td.Normal(torch.zeros(self.num_fixed_params, device=self.device),
torch.ones(1, device=self.device)/tau_alpha)
zeta_prior = td.Normal(torch.zeros(self.num_mixed_params, device=self.device),
torch.ones(1, device=self.device)/tau_zeta)
Our goal is to use Variational Inference to compute (approximate) posterior distributions over \(\boldsymbol\tau_\alpha\) and \(\boldsymbol\tau_\zeta\). Therefore, we must introduce two new approximate distributions in the compute_variational_approximation_q() method: \(q(\boldsymbol\tau_\alpha)\) and \(q(\boldsymbol\tau_\zeta)\). Will use Gamma distributions as the approximate distribution for \(\boldsymbol\tau_\alpha\) and \(\boldsymbol\tau_\zeta\).
# q(tau_alpha) - construct posterior approximation on tau_alpha
q_tau_alpha = td.Gamma(self.softplus(self.tau_alpha_alpha), self.softplus(self.tau_alpha_beta))
# q(tau_zeta) - construct posterior approximation on tau_zeta
q_tau_zeta = td.Gamma(self.softplus(self.tau_zeta_alpha), self.softplus(self.tau_zeta_beta))
Note that these 2 new variational distributions have learnable parameters (we will later optimize the ELBO w.r.t. their values) which we will have to initialize in the initialize_variational_distribution_q() function:
# q(tau_alpha) - initialize parameters of InverseGamma approximation
self.tau_alpha_alpha = nn.Parameter(-1*torch.ones(self.num_fixed_params))
self.tau_alpha_beta = nn.Parameter(-1*torch.ones(self.num_fixed_params))
# q(tau_zeta) - initialize parameters of InverseGamma approximation
self.tau_zeta_alpha = nn.Parameter(-1*torch.ones(self.num_mixed_params))
self.tau_zeta_beta = nn.Parameter(-1*torch.ones(self.num_mixed_params))
At this point, we are almost done. We are just missing updating the ELBO, which now has to include two additional KL terms that result from the two new priors in the generative process: \(\mbox{KL}[q(\boldsymbol\tau_\alpha) || p(\boldsymbol\tau_\alpha)]\) and \(\mbox{KL}[q(\boldsymbol\tau_\zeta) || p(\boldsymbol\tau_\zeta)]\). We can add these 2 extra terms to the ELBO by including the following 2 lines in the ELBO computation done in by the elbo() function:
# KL[q(tau_alpha) || p(tau_alpha)]
kld += td.kl_divergence(q_tau_alpha, tau_alpha_prior).sum()
# KL[q(tau_zeta) || p(tau_zeta)]
kld += td.kl_divergence(q_tau_zeta, tau_zeta_prior).sum()
And we are done! We have implemented ARD in the core MXL model. After convergence of the ELBO, we can inspect the values of the variational parameters self.tau_alpha_alpha, self.tau_alpha_beta, self.tau_zeta_alpha and self.tau_zeta_beta for the results of the ARD selection process as we have done above in this notebook.